💻4. Analyzing the Results
How to deal with them spreadsheets!!
So, you've extracted all that data. Now, how do we deal with those spreadsheets!
To make life easier, we've added some options to merge the spreadsheets in all those folders and subfolders after GAT analysis. To do this, go to GAT->Analysis.
Currently, there are two options:
Merge Results: Merge csv files that match the filename provided across multiple folders.
Merge Results multiple csvs: Merge multiple csvs across different folders.
What do the files mean?
For example, I've analyzed an enteric neuronal dataset from Hamnett 2022 et al.. The raw data is accessible on Zenodo. The dataset of interest is a Calbindin, Calretinin and Hu immunolabelled dataset: EXP174 (4 x distal colon (DC) and 4 x proximal colon (PC)).
After analysis, we get 8 folders like this:
├───EXP174 2022_06_15 Ca_DC1
├───EXP174 2022_06_15 Ca_DC2
├───EXP174 2022_06_15 Ca_DC3
├───EXP174 2022_06_15 Ca_DC4
├───EXP174 2022_06_15 Ca_PC1
├───EXP174 2022_06_15 Ca_PC2
├───EXP174 2022_06_15 Ca_PC3
├───EXP174 2022_06_15 Ca_PC4
During analysis, I enter DC1, PC1 etc... to distinguish each replicate and tissue region. This makes downstream analysis easier.
Within a folder, the files are organised like below (click to expand, and then click on each entry to read a description):
Example of the files within a folder
│ CalB_CalR_ROIs.zip
│ CalB_ROIs.zip
│ CalR_ROIs.zip
│ Cell_counts.csv
│ Ganglia_ROIs_MAX_EXP174 2022_06_1_PC4.zip
│ Hu_label_MAX_MAX_EXP174 2022_06_1_PC4.tif
│ Hu_ROIs_MAX_EXP174 2022_06_1_PC4.zip
│ MAX_MAX_EXP174 2022_06_1_PC4.tif
└───spatial_analysis
CalB_around_CalR.tif
CalB_around_Hu.tif
CalB_coordinates.csv
CalR_around_CalB.tif
CalR_around_Hu.tif
CalR_coordinates.csv
Hu_around_CalB.tif
Hu_around_CalR.tif
Hu_coordinates.csv
Hu_neighbours.tif
Neighbour_count_CalB_CalR.csv
Neighbour_count_Hu.csv
Neighbour_count_Hu_CalB.csv
Neighbour_count_Hu_CalR.csv
The names of the file may vary based on analysis and future updates, but the structure should remain the same.
Merge Results
As you can see, there are multiple csv files in every folder. It becomes a challenge to comb through each directory and merge them. If you were interested in the summary of cell counts, you would want to combine all the files with the name: Cell_counts.csv as one. To do this, go to GAT -> Analysis -> Merge Results as .
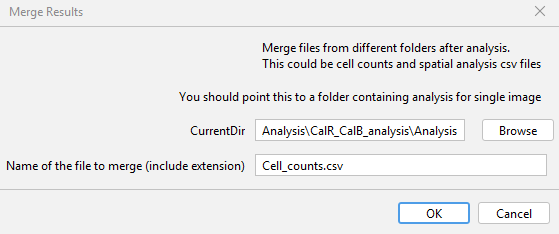
Choose the parent directory with all the analysis folders for CurrentDir. In the next row, enter the exact name of the file with the extension. As I'm interested in cell counts, I enter Cell_counts.csv. After you click ok, it will go through each directory, find the file with matching filename and then merge them into one big file. In this case, the merged file would have the name: Merged_Analysis_Cell_counts.csv in the parent directory.
Merge Results multiple csvs
This is similar to Merge Results, but the difference is it will merge all csv files. It scans the first folder and creates a list of csv files to summarise. It searches for these files across all the subsequent folders and merges them.
Data Analysis
For data analysis, I often use Orange as its an interactive and freely available software. It uses visual programming, where you drag and drop 'widgets' to create analysis workflows. Its written in Python and has loads of tutorials, both written and on Youtube. I will use Orange to demonstrate some of the analysis you could do with the Summary data from above. Knime is an example of another similar software but with way more options.
Download the software and install it on your machine.
Once installed, double click and start a New Project. I won't go into too many details on how the software works. Basically, the widgets on the left are like building blocks of a data analysis workflow. Widgets are grouped into classes according to their function. For more details, I highly recommend the introductory tutorials.
Loading the Data
As the summary data is in csv format, we drag and drop the CSV File Import widget onto the canvas

Double click on the CSV File Import widget and you can choose the file to import. Click on the folder icon to select a csv file.

Once opened, you will get an Import Options dialog with table and values. If you right click on a column, you can change the type. Column 8 is just a divider, so I've 'Ignored' that column. Otherwise, everything column with strings that can distringuish between experiments or treatments can be set as Categorical. You can also set everything to 'Auto' and see if that works too.



Once the table is imported, if you want to visualize it, you can click and drag from the CSV File Import Widget. If you release the mouse click a list of widgets will appear. Select Data Table. For more info on how to create workflows, look at this tutorial. Double clicking on the data table will reveal a table corresponding to the data imported.

Creating Classes
Now, we would like to group the data into distal and proximal regions of the colon. To do this, we use the Create Class widget. The column Experiment has experiment names, where the suffix DC for distal colon and PC for proximal colon. We use this information to create classes so data can be analyzed based on each region.

If we connect a Data Table widget to the output of Create Class, we can see a new column called Region.

Visualizing Results: Box Plot
Now, lets connect the Box Plot widget

We can visualize our results by double clicking the Box Plot widget. For example, we can compare the average number of neurons for each region by choosing Total Hu as the Variable and Region as the Subgroup.

Choosing Experiment for `Subgroups` will show Total Hu per experiment.
Number of neighbours: Distribution
Example workflow for frequency vs number of neighbours around each neuron (not normalised). Data used: Merged_Analysis_Neighbour_count_Hu.csv

Last updated