Troubleshooting
Every effort has been made to make sure the installation and configuration works across devices (Mac and Windows OS), however, there will always be some unforeseen bugs.
Make sure you follow the installation and configuration first.
Tips:
Due to recent udpated in Fiji and GAT, downloading Fiji again and reinstalling GAT from scratch will most likely fix your problem.
Please try the analysis on the sample images provided here to ensure you have installed GAT properly.
To check system requirements, go to this page on the left.
Common issues
img_resize not found -> DeepImageJ error
If you are getting errors during the model prediction steps and get a similar error as below:

It means you have not initialized DeepImageJ before running GAT. To fix it, go to Plugins -> DeepImageJ -> DeepImageJ Run. You will see a Installation progress bar on top right of the DeepImageJ window. Once this is complete, restart Fiji and try running GAT again.

Missing Model Files
If you get an error saying, Cannot finding the model file , there are two possible ways to fix it:
Update GAT again and restart Fiji
OR
Start with a fresh installation of Fiji and install GAT again.
Slow model predictions
If the cell or ganglia segmentation steps are taking longer, you could speed it up if you have a graphics card (GPU). To set it up, check this site for more info.
Pytorch 2.4.1 was used for ganglia model and Tensorflow 1.15 for the enteric neuron models
Tensorflow errors
If the images are not being segmented and you get the following or a similar output in the Console:
[WARNING] The JVM seems to have crashed when loading the TensorFlow JAR library: file:/Applications/Fiji.app/jars/libtensorflow-1.15.0.jar
[WARNING] Please run Edit > Options > TensorFlow and choose another version or delete the crash file to load the JAR library again.
[ERROR] Could not load TensorFlow. Check previous errors and warnings for details.
[WARNING] CSBDeep canceled.
you may need to change the Tensorflow version on your Fiji installation.
Follow the instructions at the CSBDeep website and try a different Tensorflow version. Make sure it has CPU on the name.
To use Tensorflow GPU you need an NVIDIA graphics card and need to install CUDA 10.1 and cuDNN>=7.5.1.
Apple Mac: M1 processor tensorflow problem
There is a known issue using GAT on Mac OS with Apple M1 chips. There are tensorflow compatbility issues when running GAT models in Fiji StarDist. Unfortunately, we do not have a fix for this. It works fine on a Mac OS with Intel chips as far as we know.
Out of memory errors
When running StarDist on really large images or if your computer has low RAM, it is possible that you may run out of memory. You can check your Task Manager in Windows or Activity monitor in Mac.
For example, you may get an error printed on your console window:

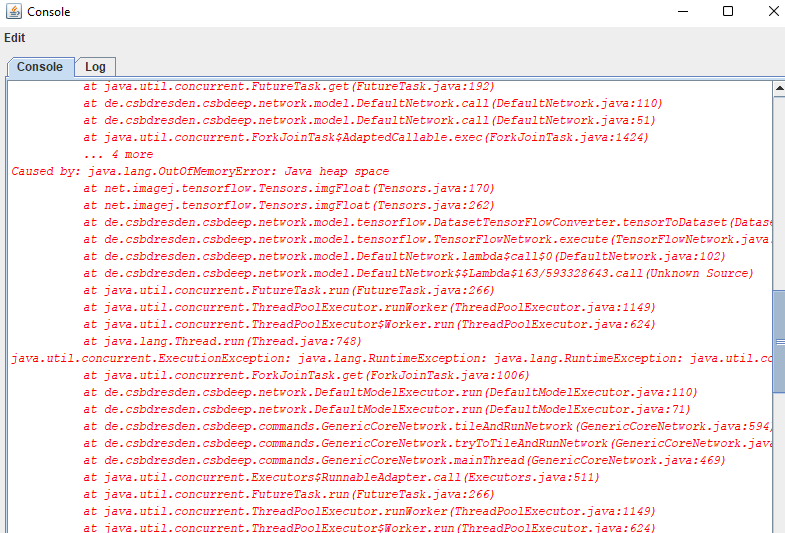
Specifically you may see: java.util.concurrent.ExecutionException: java.lang.OutOfMemoryError: Java heap space
This means you ran out of memory. If you keep getting this error and if GAT says no cells found even though there are, then you may have to either:
use a computer with more RAM (16 GB or higher, depends on image size)
use QuPath for analysis as it is more memory efficient. You could export the ROIs and use it within FIJI as well.
CLIJ errors
CLIJ is a powerful and user-friendly GPU-accelerated image processing library. GAT uses CLIJ under the hood to speed up things and future development will use GAT extensively. If you get errors related to CLIJ, please have a look at this webpage. Make sure you update your GPU drivers as well.
You will also need 3D ImageJ suite to be added to update sites.
Miscellaneous Errors
Filepath or filename related issues
If your file path contains commas or hyphens, Fiji/GAT can run into errors. This can be a sneaky one and harder to troubleshoot.
For example,
E:\C\C090 - Gene knockout day 23, test-1\Processed Images
This will cause errors during analysis and potentially stop mid way. The errors won't give you any indication that this is the issue either, which complicates it. However, it is good convention to shorten your filename, remove hyphens,commas, and use underscores instead of space. This filename could instead be
E:\C\C090_gene_KO_day23_test1\processed_images
I would recommend reading this helpful resource for better ideas on how to create filenames.
Installation issue:
Manually enter an update site
If you want to manually enter an update site after you've selected Help -> Update. Click on “Add Unlisted site”

A new row will appear with empty boxes under URL column.

You have to enter the "Name" first. So, double click on "New".

After entering the name, double click on the corresponding URL box and enter the name of the update site. Make sure there is no forward slash '/' at the end of the URL.

If its for the calcium imaging analysis, enter: https://sites.imagej.net/Template_Matching
If its for GAT, enter:
https://sites.imagej.net/GutAnalysisToolboxClick outside the URL box or press enter and if everything is well, you will get a box like this:

Model files installation error
The model files will automatically be installed from the update site. Please update ImageJ by going to Help -> Update ImageJ. Once updated, restart Fiji. If you already installed GAT, untick GAT update site and try the GAT installation process again.
The models for neuron and ganglia segmentation are within
Fiji.app/modelsfolder. They contains models for:Enteric neuron model: 2D_enteric_neuron_v4_1.zip
Enteric neuron subtype model: 2D_enteric_neuron_subtype_v4.zip
Ganglia model: 2D_Ganglia_RGB_v3.bioimage.io.model
The ganglia model was updated to be compatible with deepImageJ v3
It uses Pytorch instead of Tensorflow.
The download may take a while as the model files are large.
Support
If you have any difficulties which are not resolved above, suggestions, or find any bugs, you can use any of the following means of getting in touch:
Go to the Feedback page on the left pane for submitting issues or questions.
OR
Create a post under Issues
OR
Post the problem on the Imagesc forum and tag @pr4deepr, @GAT
Last updated