Contributing to GAT
Any contributions are welcome! This can be feedback about the software, suggestions for new analysis workflows, or even contributions to the code. Importantly, you do not need to be a programmer to contribute to GAT, as the most valuable contributions from the end users who usually tend to be non-coders.
One of the main contributions we are after are manually annotated images for generating more robust models that can segment enteric neurons, neuronal subtypes, glial cells or any other cell type within the gut wall.
Improving the segmentation models
The deep learning (DL) models on GAT have been trained on a wide variety of images so that it can work on commonly used markers (Hu, nNOS, Calbindin etc..) and modalities (widefield & confocal projections). However, these models are only as good as the images it has been trained on. It is likely GAT may not work well on some of your images, which means these would be perfect images for training and improving the GAT models.
The contributions can be for:
neuronal soma - "Hu" labelling
neuronal subtypes such as nNOS, Calbindin, Calretinin etc.. from any species, or
ganglia outlines using different combinations of a neuronal/glial marker and Hu.
There are two ways you can contribute training data:
Create training data as you analyze
Create training data using specialised scripts
Create training data as you analyze
In the latest version of GAT, you should see the following section:


You can perform analysis as you normally would and manually correct your segmentations using the ROI tools and ROI Manager. Watch this video on how to do so. If you tick the box and specify a folder location, the manual corrections will be saved in folders.


If you have only performed analysis for the neurons, you will only have the Cells folder. If you are segmenting the ganglia as well, you will also get a folder named Ganglia.
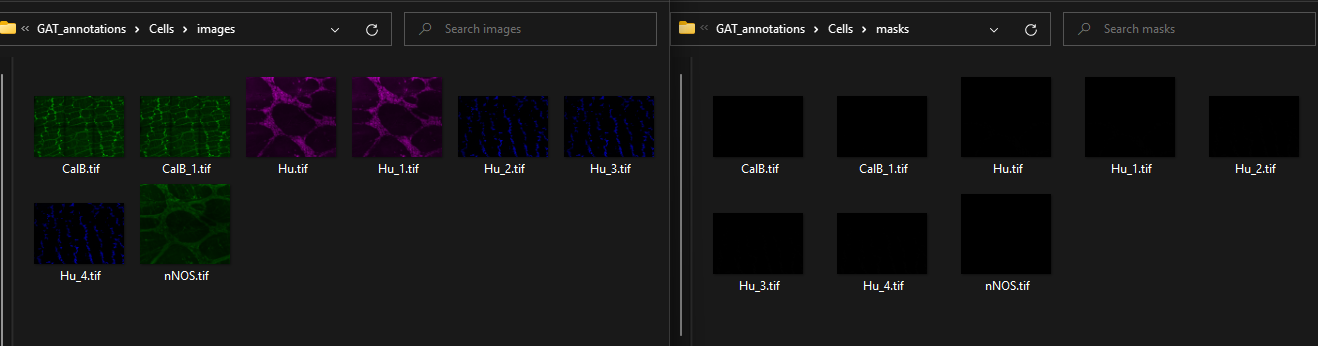
Within each folder, you will have 2 folders: images and masks:


The images folder contains the original image for the marker/s that were being analysed, and the masks folder will contain the corresponding image where each cell has an individual colour (label image).

This will be the same for the Ganglia folder, but the ganglia mask will be a binary image (black and white).
These folders with images and masks are whats needed for training. We will provide an location online to upload the images, but if you already have some, please post under the discussions or issues tab. You could also contact me at pradeep.rajasekhar@gmail.com.
It is best to get atleast 5 - 10 images of varying image and labelling qualities that are representative of the images that you encounter in your lab.
Create training data using specialised scripts
Will be updated..
Whats in it for you?:
The new model will work on your images and reduce the analysis time for images acquired at your lab. You can focus more time on writing/experiments or fun things instead of manually counting cells!
Your contributions can not only be used for GAT, but for other software developers who want to develop better analytical tools for enabling ENS research.
Improve the generalisability of the model and contribute to improving ENS research.
You will be acknowledged for your contributions.
Last updated